Data processing
We offer processing of MR data measured at our core facility or elsewhere. We use standard processing techniques provided by the scanner's software (ParaVision), external software tools (FSL, DTI Studio, ...) and software developped or co-developped by our team as a part of our research projects. Main tools:
jMRUI
jMRUI (developed by the MRUI Consortium) is a software package for advanced time-domain analysis of magnetic resonance spectroscopy (MRS) and spectroscopic imaging (MRSI) data. Through a user-friendly graphical interface biomedical researchers and clinical radiologists can use state-of-the-art signal processing algorithms to analyse and quantify spectroscopic data. A member of our team currently oversees software development and management. If you have inquiries regarding new functionality or any other requests related to MR spectra processing, please contact us at jmrui@isibrno.cz. For more information about jMRUI see website www.jmrui.eu.
 jmrui_header_image_1b-1024x625.png 267.05 KB
jmrui_header_image_1b-1024x625.png 267.05 KB
NMRScope-B
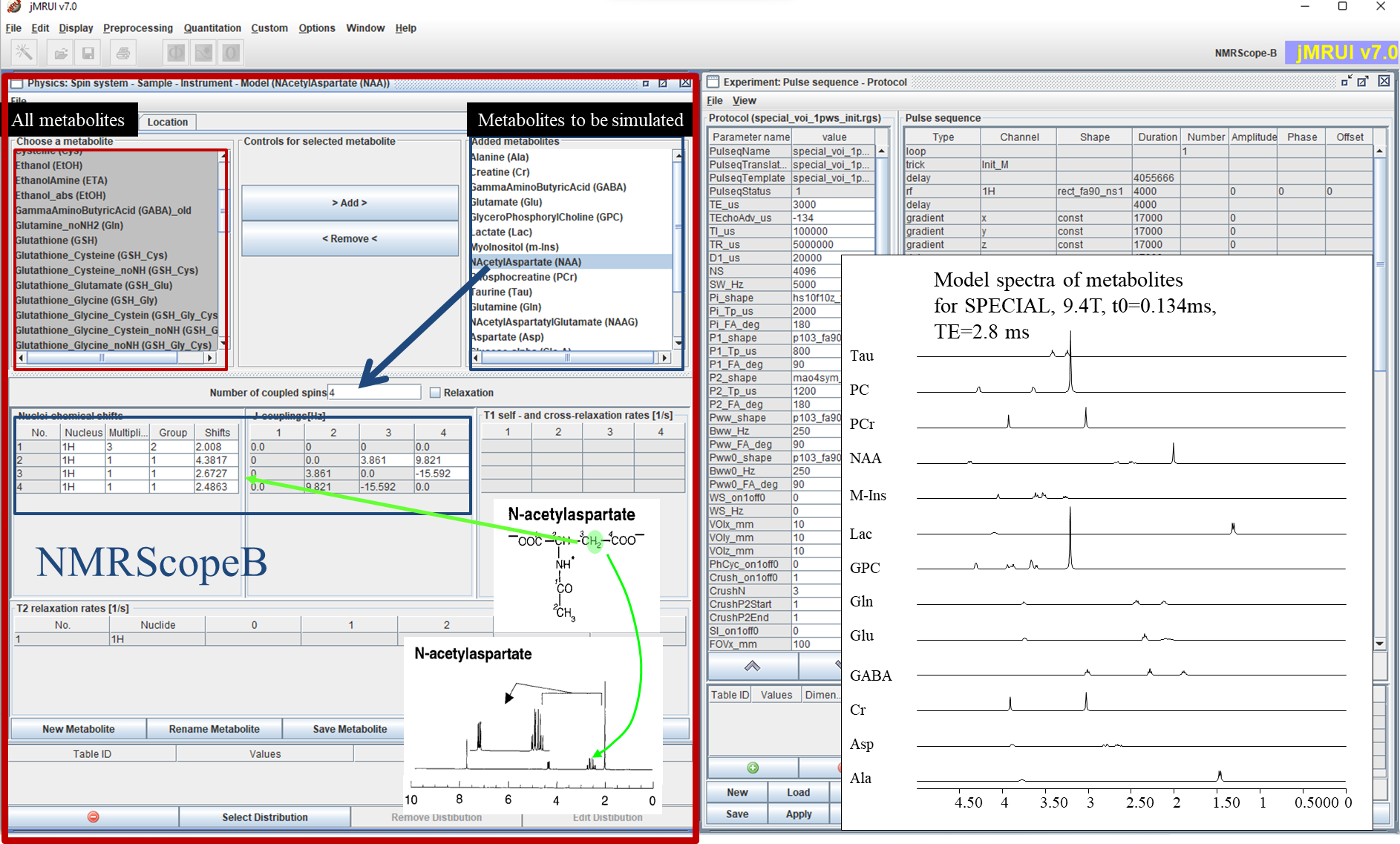
The Virtual Scanner, NMRScope-B, is a powerful simulation tool integrated into jMRUI. It allows simulation of basis sets for various sequences such as STEAM, PRESS, MEGA-PRESS, semi-LASER, or SPECIAL, which can be used in advanced fitting algorithms like, e.g., AQSES, QUEST, LCModel. The software is also designed for testing new sequence ideas, optimizing existing protocols, and for educational purposes. The NMRScope-B computational core has been developed and maintained solely by our team. If you have an inquiry, please contact us through the email jmrui@isibrno.cz. For more information see jMRUI website http://www.jmrui.eu/features/virtual-scanner/.
 NMRScopeB-servises.png 854.83 KB
NMRScopeB-servises.png 854.83 KB
PerfLab
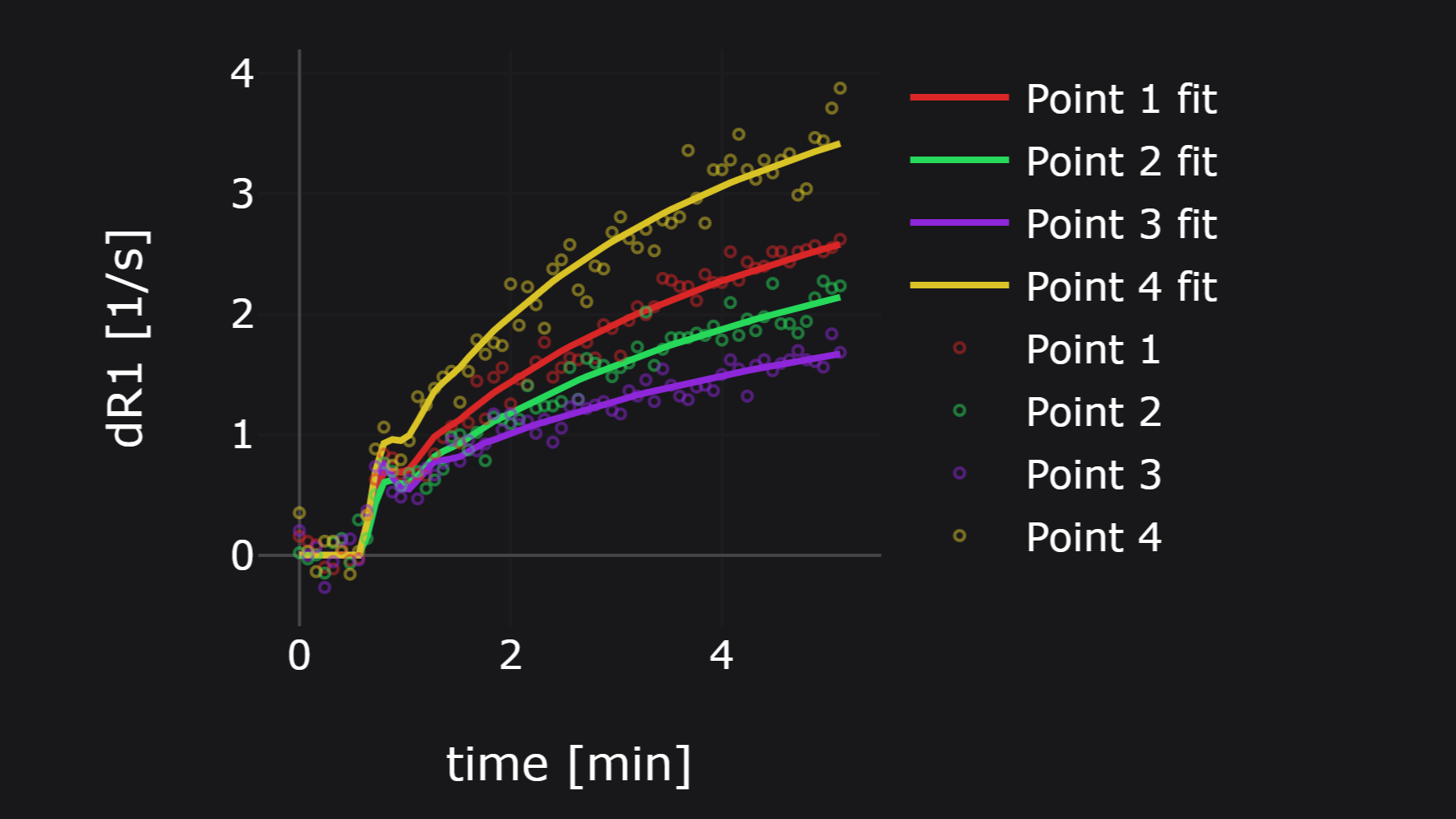
PerfLab is a web-based tool for quantitative analysis of DCE/DSC-MRI datasets. The software has been developed and maintained solely by our team. If you have any inquiry, please contact us through the email perflab@isibrno.cz. For more information see PerfLab website perflab.isibrno.cz.
DWI/DTI/DKI/IVIM Data Processing and Analysis Tools
We use a wide range of specialized software tools to process and analyze data from our DWI, DTI, DKI, and IVIM experiments. Our workflows cover everything from basic image reconstruction and artifact correction to advanced modeling of diffusion and perfusion parameters, tractography, and quantitative statistical analysis.
Depending on the type and complexity of the dataset, we utilize the following tools:
- Paravision 360 – for MRI acquisition, preprocessing, and initial reconstruction
- DSI Studio – for diffusion modeling, tractography, and visualization of fiber orientation
- FSL (FMRIB Software Library) – for diffusion tensor fitting, brain extraction, registration, and statistical analysis
- MITK Diffusion – for IVIM modeling, visualization
- ExploreDTI – for tensor and kurtosis model fitting
- ImageJ / Fiji – for general-purpose image processing and ROI analysis
- MATLAB and Python – for custom scripts, model fitting, quantitative analysis, for data handling, visualization and implementation of advanced models
- SPM (Statistical Parametric Mapping) – correction of signal inhomogeneities related to T2*
Our team is experienced in developing custom processing pipelines and integrating multi-modal datasets, ensuring high-quality results tailored to your research goals.
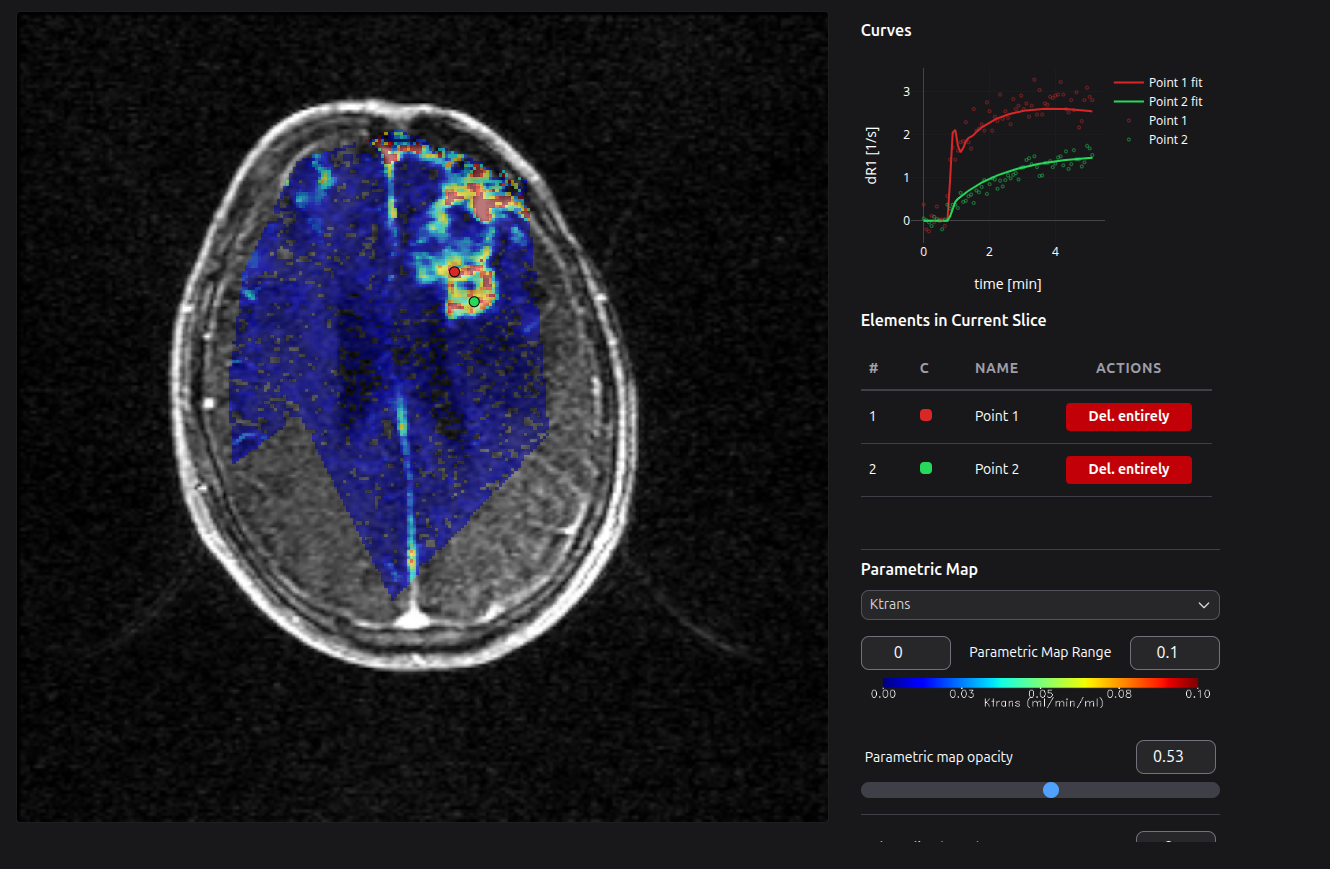 Perflab web interface
Perflab web interface